Table of Contents
 Introduction to Antibiotic Resistance
Introduction to Antibiotic Resistance
Antimicrobial compounds, including natural and synthetic antibiotics, have been crucial in combating infections. Antibiotic resistance, however, has risen rapidly, threatening public health. Resistance can develop through mutations or acquisition of resistance genes via horizontal gene transfer, which has become the primary driver of the current antimicrobial resistance pandemic.
Origins of Antibiotic Resistance
Antibiotic resistance is ancient, arising from interactions between organisms and their environment. Many antibiotic-producing bacteria, such as Streptomyces species, carry self-resistance mechanisms. Environmental non-antibiotic-producing bacteria have also evolved resistance to survive alongside these producers. Even ancient permafrost samples reveal genes resistant to β-lactams, tetracyclines, and glycopeptides, showing the long-standing presence of resistance genes.
(https://share.google/images/BHfaQxPqmbpyS3GPC)
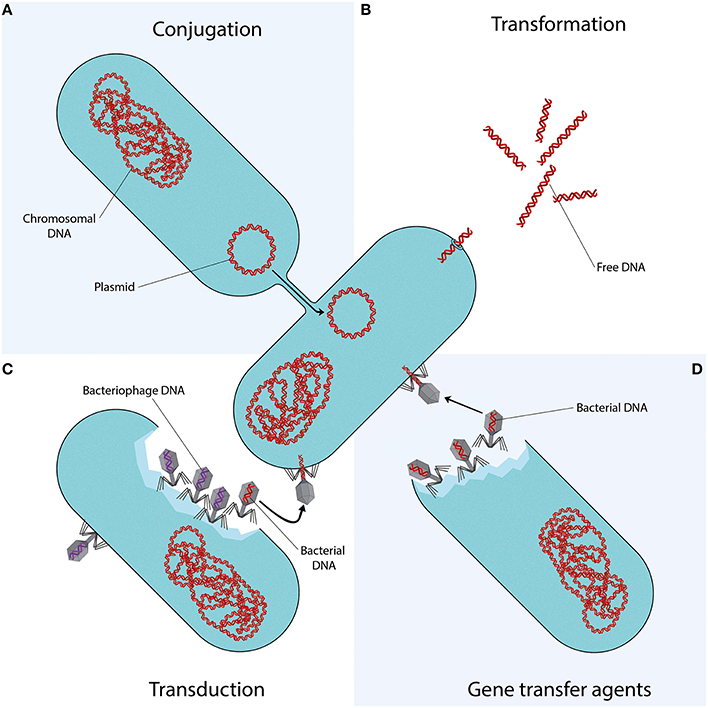
Mechanisms of Horizontal Gene Transfer
Conjugation
Conjugation involves direct transfer of DNA from one bacterium to another, often via plasmids. Plasmids carrying mobile genetic elements such as transposons or integrons spread resistance genes across bacterial populations, including clinically important genes like blaCTX-M and quinolone resistance genes.
Transformation
In transformation, bacteria uptake DNA fragments from their environment. For instance, penicillin and streptomycin resistance genes were transferred between Streptococcus pneumoniae strains in early experiments, demonstrating this mechanism’s role in spreading resistance.
Transduction
Transduction occurs when bacteriophages transfer DNA between bacteria “by accident.” This process contributes to resistance evolution in species like Staphylococcus aureus and other clinically relevant bacteria.
Environmental Factors and Antibiotic Use
Widespread antibiotic use in medicine, agriculture, and aquaculture accelerates resistance by increasing selective pressure. Most antibiotics are excreted unchanged into the environment, creating hotspots for resistance gene transfer. Increased selection pressure has also accelerated horizontal gene transfer and the abundance of resistome elements.
Conclusion
Horizontal gene transfer—including conjugation, transformation, and transduction—is key to spreading antibiotic resistance genes among bacteria. Understanding these mechanisms is critical to combating the rise of resistant pathogens and protecting public health.
To support responsible antibiotic use, Bioguard offers the miniAST Veterinary Antibiotic Susceptibility Test Analyzer, a tool designed to help combat antimicrobial resistance with game-changing features:
| Feature | Benefit |
| Fast Results | Get results in just 6 hours, enabling swift and confident treatment. |
| Automated Interpretations | Instantly deliver precise susceptibility profiles, supporting faster, more informed clinical decisions and optimizing patient care. |
| Dual-Sample Testing | Double the efficiency with simultaneous analysis of two samples at once. |
| High Accuracy | Achieve an impressive 92% accuracy rate compared to traditional disc diffusion tests. |
📌 Note for Veterinarians:
The miniAST Veterinary Antibiotic Susceptibility Test Analyzer is available exclusively to licensed veterinary clinics and hospitals.
📩 How to Order miniAST
To purchase miniAST or request a quotation, please contact our sales team or email our customer service:
📧 service@bioguardlabs.com
☎️ Please include your hospital name and contact number so our sales representative can follow up with you directly.
Source:
- Akrami F, Rajabnia M, Pournajaf A. Resistance integrons; A Mini review. Caspian J Intern Med. 2019. 10(4):370-376.
- Babakhani S, Oloomi M. Transposons: the agents of antibiotic resistance in bacteria. J Basic Microbiol. 2018. 58(11):905-917. doi: 10.1002/jobm.201800204. Epub 2018 Aug 16.
- Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Meier-Kolthoff JP, Klenk HP, Clément C, Ouhdouch Y, van Wezel GP. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol Mol Biol Rev. 2015. 80(1):1-43.
- Barlow M, Hall BG. Phylogenetic analysis shows that the OXA beta-lactamase genes have been on plasmids for millions of years. J Mol Evol. 2002. 55(3):314-21.
- Barlow M. What antimicrobial resistance has taught us about horizontal gene transfer.
- Methods Mol Biol. 2009. 532:397-411.
- Bello-López JM, Cabrero-Martínez OA, Ibáñez-Cervantes G, Hernández-Cortez C, Pelcastre-Rodríguez LI, Gonzalez-Avila LU, Castro-Escarpulli G. Horizontal Gene Transfer and Its Association with Antibiotic Resistance in the Genus Aeromonas spp. Microorganisms. 2019. 7(9).
- Cabello FC. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol. 2006. 8(7):1137-44
- Clark CA, Purins L, Kaewrakon P, Focareta T, Manning PA.
- The Vibrio cholerae O1 chromosomal integron. Microbiology. 2000. 146 ( Pt 10):2605-2612..
- Datta N, Hughes VM. Plasmids of the same Inc groups in Enterobacteria before and after the medical use of antibiotics. Nature. 198. 306(5943):616-7.
- D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006. 311(5759):374-7.
- D’Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. Antibiotic resistance is ancient. Nature. 2011. 477(7365):457-61
- Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect Drug Resist. 2015. 8:49-61.
- Graham DW, Olivares-Rieumont S, Knapp CW, Lima L, Werner D, Bowen E. Antibiotic resistance gene abundances associated with waste discharges to the Almendares River near Havana, Cuba. Environ Sci Technol. 2011. 45(2):418-24.
- Griffith F. The Significance of Pneumococcal Types. J Hyg (Lond). 1928. 827(2):113-59.
- Haaber J, Penadés JR, Ingmer H. Transfer of Antibiotic Resistance in Staphylococcus aureus. Trends Microbiol. 2017. 25(11):893-905.
- Harford N, Mergeay M. Interspecific transformation of rifampicin resistance in the genus Bacillus. Mol Gen Genet. 1973. 120(2):151-5.
- Hotchkiss RD. Transfer of penicillin resistance in pneumococci by the desoxyribonucleate derived from resistant cultures. Cold Spring Harb Symp Quant Biol. 1951;16:457-61.
- Hyder SL, Streitfeld MM. Transfer of erythromycin resistance from clinically isolated lysogenic strains of Streptococcus pyogenes via their endogenous phage. J Infect Dis. 1978. 138(3):281-6.
- Knapp CW, Dolfing J, Ehlert PA, Graham DW. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ Sci Technol. 2010. 44(2):580-7.
- Kristensen BM, Sinha S, Boyce JD, Bojesen AM, Mell JC, Redfield RJ. Natural transformation of Gallibacterium anatis. Appl Environ Microbiol. 2012. 78(14):4914-22.
- Machado E, Coque TM, Cantón R, Sousa JC, Peixe L. Antibiotic resistance integrons and extended-spectrum {beta}-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J Antimicrob Chemother. 2008. 62(2):296-302.
- Mazaheri Nezhad Fard R, Barton MD, Heuzenroeder MW. Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett Appl Microbiol. 2011. 52(6):559-64.
- Nandi S, Maurer JJ, Hofacre C, Summers AO. Gram-positive bacteria are a major reservoir of Class 1 antibiotic resistance integrons in poultry litter. Proc Natl Acad Sci U S A. 2004. 101(18):7118-22.
- Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009. 33(4):757-84.
- Ploy MC, Chainier D, Tran Thi NH, Poilane I, Cruaud P, Denis F, Collignon A, Lambert T. Integron-associated antibiotic resistance in Salmonella enterica serovar typhi from Asia. Antimicrob Agents Chemother. 2003. 47(4):1427-9.
- Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 2006. 313(5783):89-92.
- Ramos, S., Igrejas, G., Silva, N., Jones-Dias, D., Capelo-Martinez, J. L., Caniça, M., et al. First report of CTX-M producing Escherichia coli, including the new ST2526, isolated from beef cattle and sheep in Portugal. Food Control. 2013.31, 208–210.
- Robicsek, A., Jacoby, G. A., and Hooper, D. C. The worldwide emergence of plasmid-mediated quinolone resistance Lancet Infect. Dis. 2006. 6, 629– 640
- Rowe-Magnus DA, Guerout AM, Ploncard P, Dychinco B, Davies J, Mazel D. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc Natl Acad Sci U S A. 2001. 98(2):652-7.
- Sarmah AK, Meyer MT, Boxall AB. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere. 2006. 65(5):725-59.
- Varga M, Kuntová L, Pantůček R, Mašlaňová I, Růžičková V, Doškař J. Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol Lett. 2012. 332(2):146-52.
- Volkova VV, Lu Z, Besser T, Gröhn YT. Modeling the infection dynamics of bacteriophages in enteric Escherichia coli: estimating the contribution of transduction to antimicrobial gene spread. Appl Environ Microbiol. 2014. 80(14):4350-62.
- Xu H, Broersma K, Miao V, Davies J. Class 1 and class 2 integrons in multidrug-resistant gram-negative bacteria isolated from the Salmon River, British Columbia. Can J Microbiol. 2011.57(6):460-7.
- Xu Z, Li L, Shirtliff ME, Peters BM, Peng Y, Alam MJ, Yamasaki S, Shi L. First report of class 2 integron in clinical Enterococcus faecalis and class 1 integron in Enterococcus faecium in South China. Diagn Microbiol Infect Dis. 2010. 68(3):315-7.
