The Danger of Antimicrobial Resistance (AMR) in Animals
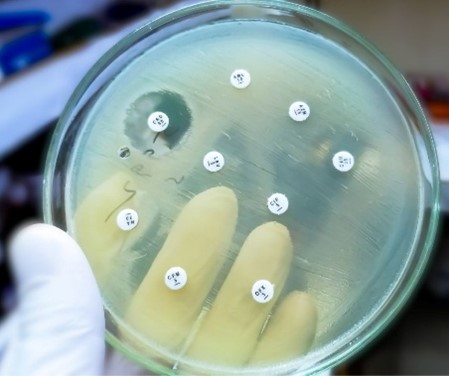
(https://share.google/images/epa3Ydpxlht3J77m3) The Invisible Pandemic Antimicrobial resistance (AMR) is now widely recognized as one of the most formidable global health threats of the 21st century. It fundamentally undermines the efficacy of existing antibiotics and jeopardizes the very foundations of modern medicine — from routine surgical safety to cancer chemotherapy and neonatal care. The relentless rise of resistance has transformed once-treatable bacterial infections into persistent, sometimes untreatable, conditions, eroding decades of medical progress. This growing crisis is fueled by the uncontrolled and often unjustified use of antimicrobial agents across human healthcare, veterinary practice, and agriculture. Without coordinated global action, AMR could evolve into the next true pandemic. The gravity of this issue is underscored by the World Economic Forum, which classifies antibiotic resistance as a transnational risk exceeding the capacity of any single organization or nation to manage alone. The global outlook is dire. Projections indicate that annual mortality linked to resistant infections may exceed 10 million deaths by 2050, far surpassing the current estimated 700,000 deaths each year, underscoring the urgent need to decode the biological mechanisms that drive this phenomenon, including how resistance arises, adapts, and moves across species and environments (O’Neill, 2016). At its core, AMR is an evolutionary challenge. The emergence and spread of resistance stem from genetic variation within bacterial populations and the selective pressures exerted by antibiotic exposure. Resistance traits can originate de novo, through spontaneous mutations or structural rearrangements during DNA replication, and subsequently proliferate via horizontal gene transfer (HGT) — the lateral exchange of genetic material between unrelated bacterial cells. HGT serves as a conduit for resistance genes to traverse species boundaries, enabling pathogens to acquire multi-drug resistance in a single event. This mechanism is particularly dominant among Gram-negative bacteria, where mobile genetic elements facilitate rapid adaptation. Ultimately, the dissemination of resistance forms a complex ecological network linking human health, animal husbandry, and environmental reservoirs such as water, food, and sewage systems. Understanding these interconnections is essential to curbing the invisible pandemic of antimicrobial resistance before it surpasses our collective capacity to respond. 1.The Genetic Arms Race — How Bacteria Learn to Resist Initiation of Resistance: Random Mutations and Natural Selection At the evolutionary level, AMR emerges from the interplay between genetic diversity and selective pressure. Random mutations introduce variation, while antibiotics act as the environmental filter determining which variants survive. Random Mutations: De Novo Innovation Resistance can originate spontaneously through replication or repair errors in bacterial DNA. These heritable mutations are passed vertically to daughter cells. Although most mutations are harmful, a rare few confer survival benefits under antibiotic stress. Bacteria’s rapid reproduction amplifies this effect: Escherichia coli can divide every 20 minutes, producing over 68 billion descendants in 12 hours. This massive population size raises the probability that resistant mutants arise purely by chance. The spontaneous mutation frequency for antibiotic resistance typically falls between 10⁻⁸ and 10⁻⁹ per generation—so in a population of 10⁹ cells, at least one resistant clone is likely to appear. Under oxidative or chemical stress, this rate may increase dramatically. When antibiotics enter the environment, these mutations become the substrate of natural selection, allowing resistant lineages to thrive while susceptible ones vanish. Natural Selection: The Filter of Survival Antibiotics impose a strong selective pressure on microbial populations. Resistant bacteria, whether by mutation or gene acquisition, enjoy a survival advantage and replicate preferentially. Over successive generations, this produces a predominantly resistant community. Resistance often entails a fitness cost, such as slower growth, yet under continuous exposure, the advantage of drug survival outweighs that penalty. Secondary “compensatory” mutations can subsequently restore growth efficiency, cementing resistant strains as dominant even in the absence of antibiotics. Examples of Resistance Initiation Mycobacterium tuberculosis — Multidrug-Resistant TB (MDR-TB) Resistance in M. tuberculosis arises almost exclusively via chromosomal mutations. For instance, rifampicin resistance results from point mutations in the rpoB gene encoding the β-subunit of RNA polymerase, which reduces drug binding. Progressive accumulation of such mutations can transform a susceptible infection into a pan-resistant strain. Staphylococcus aureus — MRSA Evolution Resistance to penicillin G emerged in S. aureus as early as 1943, within a year of its introduction. Later, the acquisition of the staphylococcal cassette chromosome mec (SCCmec)—a mobile element conferring methicillin resistance—produced methicillin-resistant S. aureus (MRSA). This element imposed minimal fitness costs, enabling pandemic clones such as ST8:USA300 to thrive. S. aureus can also acquire rifampicin resistance through independent rpoB mutations, exemplifying the synergy of mutation and horizontal gene transfer. Escherichia coli — Multidrug-Resistant Strains MDR E. coli often combine chromosomal and plasmid-borne mechanisms. Mutations in gyrA, gyrB, and parC yield fluoroquinolone resistance, while plasmids carrying CTX-M β-lactamases confer cephalosporin resistance. The pandemic clone ST131 illustrates this convergence, integrating both pathways to achieve broad resistance. Environmental surveillance reveals such strains in wastewater systems, highlighting how human waste streams perpetuate environmental dissemination. Selective Pressure from Antibiotic Overuse The misuse of antibiotics across medicine, agriculture, and aquaculture accelerates resistance evolution by continuously applying selective pressure. In Healthcare Antibiotics are indispensable but frequently misused. Inappropriate prescribing and patient demand sustain excessive use. Resistant mutants can dominate within days of therapy initiation. Hospitals, intensive care units, and nursing facilities become reservoirs of entrenched resistance. In Agriculture and Food Production The agricultural sector remains a powerful amplifier of AMR. Antibiotics are administered to healthy livestock for prophylaxis and growth promotion, and used in crops and aquaculture. This continuous exposure fosters resistance transfer between animals, humans, and the environment. The colistin resistance gene mcr-1, first detected in China, arose from decades of agricultural colistin use and has since spread worldwide. Food products themselves act as vehicles for resistant bacteria, underscoring the importance of a One Health approach that unites human, animal, and environmental management. Visual Aid — Mutation-Driven vs. Acquired Resistance Mechanism Description Example Pathogen Clinical Impact Mutation (Vertical Transmission) Random chromosomal alterations that modify target structures (e.g., ribosomal subunits, enzymes). Mycobacterium tuberculosis Drives multidrug- and pan-resistant TB; also mediates fluoroquinolone resistance via gyrA/gyrB mutations. Gene Acquisition (Horizontal Transfer) Uptake
